Introducing LigoSpace: Advancing Bioactivity Prediction at NeurIPS 2025

We’re excited to share that our latest research, LigoSpace, developed in collaboration with Nanyang Technological University Singapore (NTU), has been presented at NeurIPS 2025, one of the world’s most prestigious conferences in AI and machine learning.
LigoSpace addresses a persistent challenge at the core of AI-driven drug discovery: How can we accurately predict the bioactivity of candidate ligands?
Why Bioactivity Prediction Is Still Hard
Ligands and endogenous substrates often compete for the same protein binding sites. A ligand’s ability to modulate protein function depends not just on how strongly it binds, but how effectively it occupies the binding pocket.
However, most existing computational methods focus only on the local interactions between protein and ligand. They rarely consider the spatial emptiness inside the pocket, the unoccupied regions that may still allow endogenous molecules to interfere.
This missing perspective means that even ligands with strong binding affinity may have weaker-than-expected biological effects if too much empty space remains.
Introducing LigoSpace
LigoSpace is designed to overcome these limitations by modeling the binding pocket with new geometric insights. It brings together three core innovations:
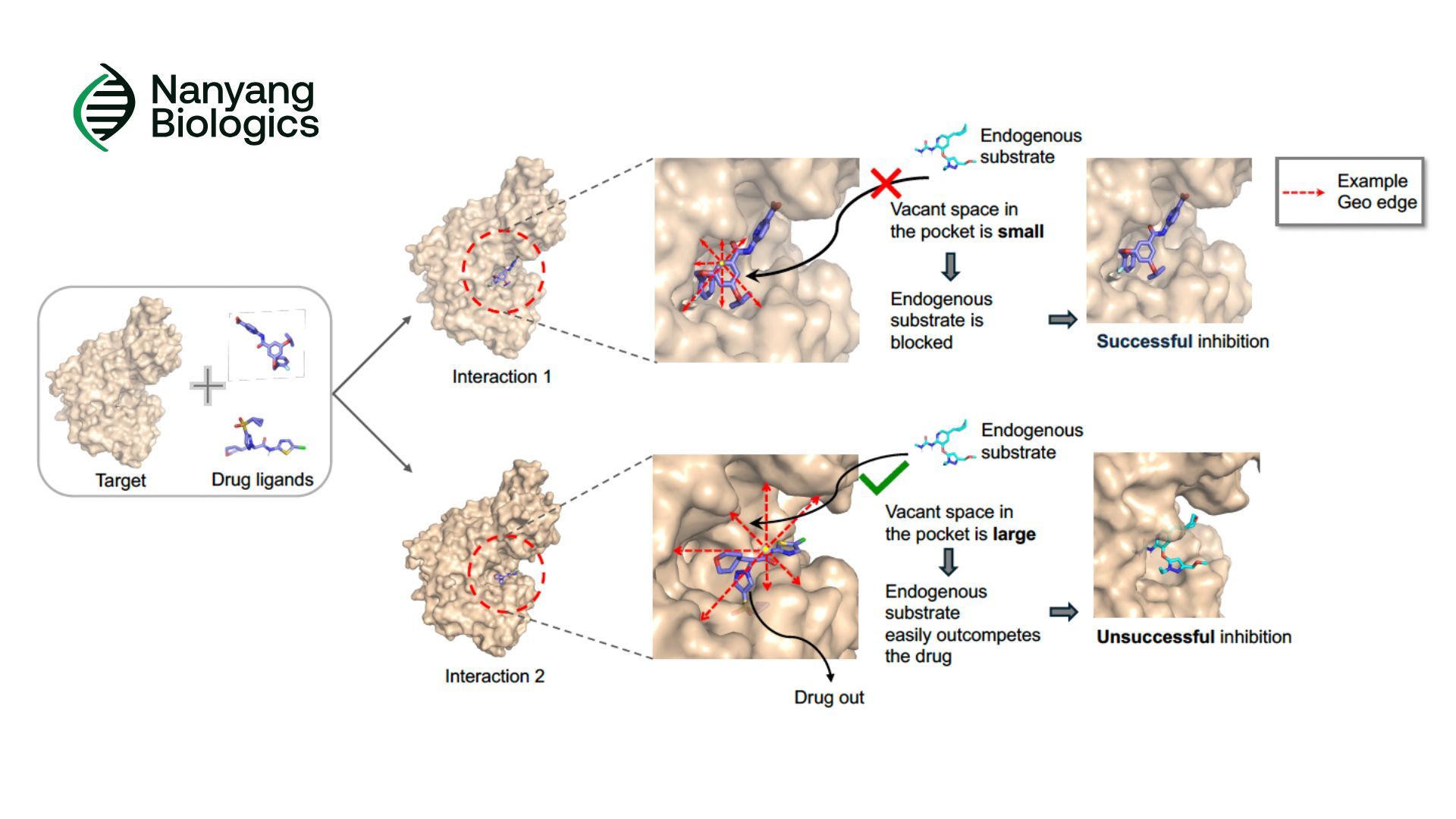
1. GeoREC: Quantifying Spatial Emptiness
GeoREC (Geometric Representation of Spatial Emptiness in Complexes) captures and encodes atomic-level empty space within protein-ligand pockets. This gives models a far more realistic understanding of how “well-occupied” a pocket truly is.
2. UnionPocket: A Unified Global Pocket View
Protein pockets are often analyzed in isolation, leading to inconsistent interpretations.
UnionPocket unifies multiple pockets into a single global representation, enabling consistency and comparability across complexes.
3. Pairwise Loss for More Meaningful Predictions
Instead of relying on conventional MSE loss, LigoSpace employs a pairwise loss function.
This better captures relative differences, an essential part of ranking ligands and prioritizing candidates in drug discovery workflows.
Learn More
- Paper: Open Review
- Poster: NeurIPS Virtual
Related Articles



